AlphaProteo Develops Innovative Proteins for Biological and Health Research
AlphaProteo: A Breakthrough in Protein Design Using AI
Introduction to AlphaProteo
Recent developments in artificial intelligence have led to the creation of AlphaProteo, a revolutionary AI system designed for protein design. This tool aims to generate proteins that can effectively bind to specific target molecules, which has significant implications for drug development, disease understanding, and various scientific research areas.
The Importance of Protein Interactions
Proteins are essential molecules in biological systems, acting as catalysts, structural components, and regulators of cellular processes. Their ability to interact with other proteins is crucial for various bodily functions, including immune responses and cell growth. Understanding how proteins bind to each other helps advance research in medical treatments and even agricultural practices.
Current Challenges in Protein Design
Although protein structure prediction tools like AlphaFold have enhanced our knowledge of protein interactions, the design of new proteins is a complex task. Traditional methods for creating protein binders involve multiple rounds of lab experiments, which are time-consuming and labor-intensive. Researchers often need to conduct various trials to optimize the binding affinity of the proteins they create.
How AlphaProteo Works
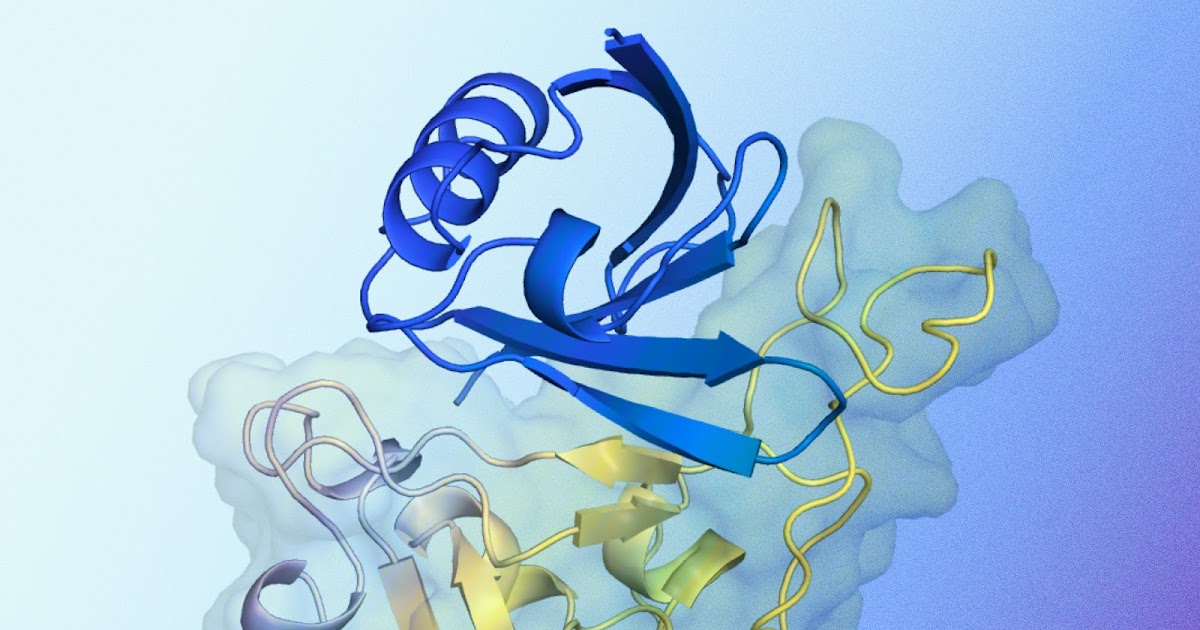
AlphaProteo changes the game by using extensive data from the Protein Data Bank and AlphaFold’s predicted structures to learn how molecules bind together. By providing AlphaProteo with the structure of a target protein and potential binding sites, researchers can generate candidates for new protein binders rapidly. This AI system boasts a higher success rate and superior binding affinities compared to existing methods.
Performance and Success Rates
AlphaProteo has demonstrated remarkable success in designing protein binders for significant targets. For instance, it has effectively created binders for:
- Viral Proteins: Such as BHRF1 and the SARS-CoV-2 spike protein receptor-binding domain.
- Cancer-Related Proteins: Including VEGF-A and PD-L1.
Scientific tests indicated that AlphaProteo’s generated candidates displayed binding strengths 3 to 300 times better than those designed through traditional methods.
Validation of AlphaProteo
AlphaProteo’s capabilities have been validated through numerous experiments conducted in collaboration with research teams at the Francis Crick Institute. For instance, an impressive 88% success rate was recorded in binding tests for the viral protein BHRF1. Furthermore, the bindings exhibited desirable biological functions, like preventing SARS-CoV-2 from infecting cells.
Exceptional Binding Capabilities
AlphaProteo has outperformed other design methods, as seen in the experimental outcomes. For example, its candidates for the TrkA protein showed even greater strength than the top prior designs, which had undergone extensive optimization. The current AI system offers optimized solutions more efficiently than previous techniques, reducing the need for extensive testing.
Future Directions for AlphaProteo
As promising as AlphaProteo appears, it has limitations; for example, it struggled to design successful binders for the target protein TNFα, which is associated with autoimmune diseases. Nevertheless, ongoing improvements are being made to expand its capabilities. The developers of AlphaProteo aim to refine the tool and broaden its application scope, exploring its potential for drug design and various other biological problems.
Responsible Development and Biosecurity
As AI continues to transform protein design, ethical considerations and biosecurity remain paramount. The researchers are taking a phased approach to share findings responsibly and engage with the scientific community to ensure the technology is developed safely. Initiatives like the NTI’s AI Bio Forum are being leveraged to develop best practices.
Opportunities for Collaboration
The team behind AlphaProteo is interested in collaborating with biologists whose research could benefit from specific protein binding. Interested parties can register for testing opportunities, contributing to the evolution of this groundbreaking technology.
Acknowledgements
This research initiative owes its success to the collaborative efforts of the Protein Design and Wet Lab teams, along with invaluable contributions from research groups at the Francis Crick Institute. The ongoing advancements in AI-driven protein design suggest a bright future for both scientific research and practical applications in health and biomedicine.






